EES 211 2.0 Data Analysis and Modeling
(Department of Physics, USJ)
Dr Thiyanga Talagala
Department of Statistics, University of Sri Jayewardenepura
What is Statistics?
What is Statistics?
The science of collecting, analyzing, presenting, and interpreting data.
What is Statistics?
The science of collecting, analyzing, presenting, and interpreting data.

Tabular data

Tabular data

Image data
Tabular data

Audio data
Image data

Tabular data

Audio data

Image data

Video data
What is Statistics?
The science of collecting, analyzing, presenting, and interpreting data.
What is Statistics?
The science of collecting, analyzing, presenting, and interpreting data.
Data analysis
ID Gender A B C Weight1 1 Male 80.0 3.6 2.5 40002 2 Female 90.0 2.5 6.3 50003 3 Female 110.0 4.0 4.5 60004 4 Female 100.0 4.5 3.2 70005 5 Female 91.5 3.0 3.5 75506 6 Male 92.0 3.9 3.7 45007 7 Male 88.0 4.2 3.8 33758 8 Male 70.0 4.6 3.9 55009 9 Female 100.5 2.9 1.2 297510 10 Female 99.8 3.7 4.2 378411 11 Female 101.5 2.7 2.2 276612 12 Female 99.2 5.1 4.4 519613 13 Female 99.6 3.8 3.9 392814 14 Female 99.1 2.7 3.1 277915 15 Female 99.8 4.0 4.8 408616 16 Female 100.4 4.2 2.8 433817 17 Female 99.3 4.1 2.9 417518 18 Female 100.8 3.2 2.5 329519 19 Female 98.8 5.0 2.8 511120 20 Female 99.0 3.9 4.4 399021 21 Female 101.4 2.9 3.9 297822 22 Female 99.0 1.3 4.5 138723 23 Female 100.4 4.6 3.1 472624 24 Female 99.6 3.5 3.9 355625 25 Female 100.4 3.5 3.7 358426 26 Female 101.7 4.4 2.1 454627 27 Female 101.6 4.3 5.3 442128 28 Female 99.7 4.1 3.6 419329 29 Female 97.7 4.4 4.3 452030 30 Female 102.5 4.3 4.5 438131 31 Female 100.7 3.6 3.4 367432 32 Female 100.5 1.5 3.2 161033 33 Female 100.0 4.1 4.4 422034 34 Female 100.5 3.4 2.5 354435 35 Female 99.8 3.3 5.5 344536 36 Female 100.4 2.0 3.1 212937 37 Female 99.6 3.0 5.2 312138 38 Female 98.6 3.9 5.0 401939 39 Female 101.0 4.9 3.6 495840 40 Female 101.5 3.4 4.1 349741 41 Female 99.7 3.9 2.5 398842 42 Female 98.7 3.4 3.8 354743 43 Female 100.6 2.1 4.5 222344 44 Female 100.0 3.1 3.6 318545 45 Female 98.3 3.1 3.0 320546 46 Female 100.0 3.4 2.8 354047 47 Female 99.4 4.6 3.5 470048 48 Female 99.7 4.3 4.6 436349 49 Female 98.8 3.3 3.0 343650 50 Female 101.8 3.2 3.4 334551 51 Female 99.7 4.2 2.2 429752 52 Female 98.4 4.1 4.0 415553 53 Female 100.2 2.8 4.8 291154 54 Female 100.3 2.8 5.0 289255 55 Male 99.0 3.4 4.3 514656 56 Male 97.1 3.8 1.6 575357 57 Male 99.4 2.9 4.0 443058 58 Male 100.6 3.9 4.0 592359 59 Male 99.9 3.4 3.1 519560 60 Male 99.9 2.4 3.7 368161 61 Male 100.6 3.3 2.6 511162 62 Male 98.8 1.9 4.2 290563 63 Male 101.1 4.4 3.2 675264 64 Male 100.0 5.0 1.9 757165 65 Male 100.7 2.6 3.1 404866 66 Male 101.0 2.0 4.9 303267 67 Male 100.2 3.6 3.2 545568 68 Male 99.1 2.9 4.2 439769 69 Male 101.2 5.4 4.4 820270 70 Male 98.0 3.0 3.5 454071 71 Male 99.5 3.7 3.1 563372 72 Male 99.7 3.0 3.0 464173 73 Male 99.8 2.3 4.2 348674 74 Male 101.0 3.2 2.4 488375 75 Male 100.1 1.2 3.7 189176 76 Male 100.4 4.5 3.2 680077 77 Male 99.9 3.2 1.2 483078 78 Male 99.8 5.2 2.1 785979 79 Male 100.7 3.5 4.4 531480 80 Male 101.1 2.3 3.3 353681 81 Male 97.6 3.6 4.3 551582 82 Male 100.6 2.1 5.4 320183 83 Male 100.4 1.7 5.0 271984 84 Male 99.6 3.3 4.2 503685 85 Male 101.0 2.6 3.9 393586 86 Male 99.6 3.0 3.3 460287 87 Male 99.7 3.1 5.1 471488 88 Male 100.9 2.4 4.1 371689 89 Male 101.7 2.4 2.3 374790 90 Male 100.3 2.9 3.3 439791 91 Male 99.6 4.2 1.6 636792 92 Male 98.8 1.5 3.3 231593 93 Male 99.7 3.6 0.9 549294 94 Male 99.1 3.3 4.8 510295 95 Male 99.7 4.1 2.9 619696 96 Male 100.4 2.7 3.1 414597 97 Male 99.1 3.4 3.3 515498 98 Male 102.6 3.3 4.1 500299 99 Male 100.2 2.5 4.2 3786100 100 Male 101.1 4.2 4.1 6410 ID Gender A B C Weight1 1 Male 80.0 3.6 2.5 40002 2 Female 90.0 2.5 6.3 50003 3 Female 110.0 4.0 4.5 60004 4 Female 100.0 4.5 3.2 70005 5 Female 91.5 3.0 3.5 75506 6 Male 92.0 3.9 3.7 45007 7 Male 88.0 4.2 3.8 33758 8 Male 70.0 4.6 3.9 55009 9 Female 100.5 2.9 1.2 297510 10 Female 99.8 3.7 4.2 378411 11 Female 101.5 2.7 2.2 276612 12 Female 99.2 5.1 4.4 519613 13 Female 99.6 3.8 3.9 392814 14 Female 99.1 2.7 3.1 277915 15 Female 99.8 4.0 4.8 408616 16 Female 100.4 4.2 2.8 433817 17 Female 99.3 4.1 2.9 417518 18 Female 100.8 3.2 2.5 329519 19 Female 98.8 5.0 2.8 511120 20 Female 99.0 3.9 4.4 399021 21 Female 101.4 2.9 3.9 297822 22 Female 99.0 1.3 4.5 138723 23 Female 100.4 4.6 3.1 472624 24 Female 99.6 3.5 3.9 355625 25 Female 100.4 3.5 3.7 358426 26 Female 101.7 4.4 2.1 454627 27 Female 101.6 4.3 5.3 442128 28 Female 99.7 4.1 3.6 419329 29 Female 97.7 4.4 4.3 452030 30 Female 102.5 4.3 4.5 438131 31 Female 100.7 3.6 3.4 367432 32 Female 100.5 1.5 3.2 161033 33 Female 100.0 4.1 4.4 422034 34 Female 100.5 3.4 2.5 354435 35 Female 99.8 3.3 5.5 344536 36 Female 100.4 2.0 3.1 212937 37 Female 99.6 3.0 5.2 312138 38 Female 98.6 3.9 5.0 401939 39 Female 101.0 4.9 3.6 495840 40 Female 101.5 3.4 4.1 349741 41 Female 99.7 3.9 2.5 398842 42 Female 98.7 3.4 3.8 354743 43 Female 100.6 2.1 4.5 222344 44 Female 100.0 3.1 3.6 318545 45 Female 98.3 3.1 3.0 320546 46 Female 100.0 3.4 2.8 354047 47 Female 99.4 4.6 3.5 470048 48 Female 99.7 4.3 4.6 436349 49 Female 98.8 3.3 3.0 343650 50 Female 101.8 3.2 3.4 334551 51 Female 99.7 4.2 2.2 429752 52 Female 98.4 4.1 4.0 415553 53 Female 100.2 2.8 4.8 291154 54 Female 100.3 2.8 5.0 289255 55 Male 99.0 3.4 4.3 514656 56 Male 97.1 3.8 1.6 575357 57 Male 99.4 2.9 4.0 443058 58 Male 100.6 3.9 4.0 592359 59 Male 99.9 3.4 3.1 519560 60 Male 99.9 2.4 3.7 368161 61 Male 100.6 3.3 2.6 511162 62 Male 98.8 1.9 4.2 290563 63 Male 101.1 4.4 3.2 675264 64 Male 100.0 5.0 1.9 757165 65 Male 100.7 2.6 3.1 404866 66 Male 101.0 2.0 4.9 303267 67 Male 100.2 3.6 3.2 545568 68 Male 99.1 2.9 4.2 439769 69 Male 101.2 5.4 4.4 820270 70 Male 98.0 3.0 3.5 454071 71 Male 99.5 3.7 3.1 563372 72 Male 99.7 3.0 3.0 464173 73 Male 99.8 2.3 4.2 348674 74 Male 101.0 3.2 2.4 488375 75 Male 100.1 1.2 3.7 189176 76 Male 100.4 4.5 3.2 680077 77 Male 99.9 3.2 1.2 483078 78 Male 99.8 5.2 2.1 785979 79 Male 100.7 3.5 4.4 531480 80 Male 101.1 2.3 3.3 353681 81 Male 97.6 3.6 4.3 551582 82 Male 100.6 2.1 5.4 320183 83 Male 100.4 1.7 5.0 271984 84 Male 99.6 3.3 4.2 503685 85 Male 101.0 2.6 3.9 393586 86 Male 99.6 3.0 3.3 460287 87 Male 99.7 3.1 5.1 471488 88 Male 100.9 2.4 4.1 371689 89 Male 101.7 2.4 2.3 374790 90 Male 100.3 2.9 3.3 439791 91 Male 99.6 4.2 1.6 636792 92 Male 98.8 1.5 3.3 231593 93 Male 99.7 3.6 0.9 549294 94 Male 99.1 3.3 4.8 510295 95 Male 99.7 4.1 2.9 619696 96 Male 100.4 2.7 3.1 414597 97 Male 99.1 3.4 3.3 515498 98 Male 102.6 3.3 4.1 500299 99 Male 100.2 2.5 4.2 3786100 100 Male 101.1 4.2 4.1 6410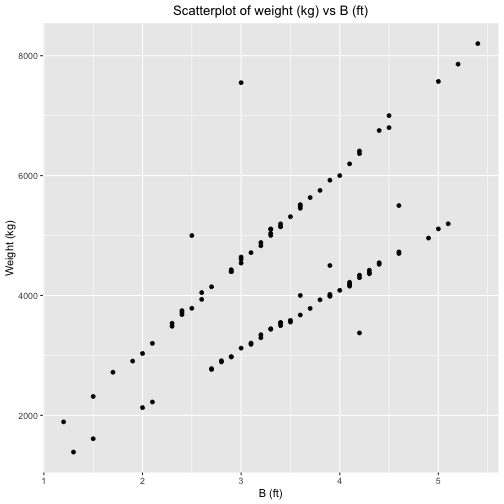
ID Gender A B C Weight1 1 Male 80.0 3.6 2.5 40002 2 Female 90.0 2.5 6.3 50003 3 Female 110.0 4.0 4.5 60004 4 Female 100.0 4.5 3.2 70005 5 Female 91.5 3.0 3.5 75506 6 Male 92.0 3.9 3.7 45007 7 Male 88.0 4.2 3.8 33758 8 Male 70.0 4.6 3.9 55009 9 Female 100.5 2.9 1.2 297510 10 Female 99.8 3.7 4.2 378411 11 Female 101.5 2.7 2.2 276612 12 Female 99.2 5.1 4.4 519613 13 Female 99.6 3.8 3.9 392814 14 Female 99.1 2.7 3.1 277915 15 Female 99.8 4.0 4.8 408616 16 Female 100.4 4.2 2.8 433817 17 Female 99.3 4.1 2.9 417518 18 Female 100.8 3.2 2.5 329519 19 Female 98.8 5.0 2.8 511120 20 Female 99.0 3.9 4.4 399021 21 Female 101.4 2.9 3.9 297822 22 Female 99.0 1.3 4.5 138723 23 Female 100.4 4.6 3.1 472624 24 Female 99.6 3.5 3.9 355625 25 Female 100.4 3.5 3.7 358426 26 Female 101.7 4.4 2.1 454627 27 Female 101.6 4.3 5.3 442128 28 Female 99.7 4.1 3.6 419329 29 Female 97.7 4.4 4.3 452030 30 Female 102.5 4.3 4.5 438131 31 Female 100.7 3.6 3.4 367432 32 Female 100.5 1.5 3.2 161033 33 Female 100.0 4.1 4.4 422034 34 Female 100.5 3.4 2.5 354435 35 Female 99.8 3.3 5.5 344536 36 Female 100.4 2.0 3.1 212937 37 Female 99.6 3.0 5.2 312138 38 Female 98.6 3.9 5.0 401939 39 Female 101.0 4.9 3.6 495840 40 Female 101.5 3.4 4.1 349741 41 Female 99.7 3.9 2.5 398842 42 Female 98.7 3.4 3.8 354743 43 Female 100.6 2.1 4.5 222344 44 Female 100.0 3.1 3.6 318545 45 Female 98.3 3.1 3.0 320546 46 Female 100.0 3.4 2.8 354047 47 Female 99.4 4.6 3.5 470048 48 Female 99.7 4.3 4.6 436349 49 Female 98.8 3.3 3.0 343650 50 Female 101.8 3.2 3.4 334551 51 Female 99.7 4.2 2.2 429752 52 Female 98.4 4.1 4.0 415553 53 Female 100.2 2.8 4.8 291154 54 Female 100.3 2.8 5.0 289255 55 Male 99.0 3.4 4.3 514656 56 Male 97.1 3.8 1.6 575357 57 Male 99.4 2.9 4.0 443058 58 Male 100.6 3.9 4.0 592359 59 Male 99.9 3.4 3.1 519560 60 Male 99.9 2.4 3.7 368161 61 Male 100.6 3.3 2.6 511162 62 Male 98.8 1.9 4.2 290563 63 Male 101.1 4.4 3.2 675264 64 Male 100.0 5.0 1.9 757165 65 Male 100.7 2.6 3.1 404866 66 Male 101.0 2.0 4.9 303267 67 Male 100.2 3.6 3.2 545568 68 Male 99.1 2.9 4.2 439769 69 Male 101.2 5.4 4.4 820270 70 Male 98.0 3.0 3.5 454071 71 Male 99.5 3.7 3.1 563372 72 Male 99.7 3.0 3.0 464173 73 Male 99.8 2.3 4.2 348674 74 Male 101.0 3.2 2.4 488375 75 Male 100.1 1.2 3.7 189176 76 Male 100.4 4.5 3.2 680077 77 Male 99.9 3.2 1.2 483078 78 Male 99.8 5.2 2.1 785979 79 Male 100.7 3.5 4.4 531480 80 Male 101.1 2.3 3.3 353681 81 Male 97.6 3.6 4.3 551582 82 Male 100.6 2.1 5.4 320183 83 Male 100.4 1.7 5.0 271984 84 Male 99.6 3.3 4.2 503685 85 Male 101.0 2.6 3.9 393586 86 Male 99.6 3.0 3.3 460287 87 Male 99.7 3.1 5.1 471488 88 Male 100.9 2.4 4.1 371689 89 Male 101.7 2.4 2.3 374790 90 Male 100.3 2.9 3.3 439791 91 Male 99.6 4.2 1.6 636792 92 Male 98.8 1.5 3.3 231593 93 Male 99.7 3.6 0.9 549294 94 Male 99.1 3.3 4.8 510295 95 Male 99.7 4.1 2.9 619696 96 Male 100.4 2.7 3.1 414597 97 Male 99.1 3.4 3.3 515498 98 Male 102.6 3.3 4.1 500299 99 Male 100.2 2.5 4.2 3786100 100 Male 101.1 4.2 4.1 6410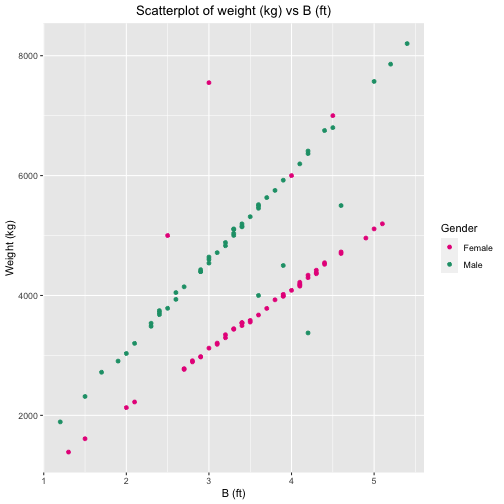
ID Gender A B C Weight1 1 Male 80.0 3.6 2.5 40002 2 Female 90.0 2.5 6.3 50003 3 Female 110.0 4.0 4.5 60004 4 Female 100.0 4.5 3.2 70005 5 Female 91.5 3.0 3.5 75506 6 Male 92.0 3.9 3.7 45007 7 Male 88.0 4.2 3.8 33758 8 Male 70.0 4.6 3.9 55009 9 Female 100.5 2.9 1.2 297510 10 Female 99.8 3.7 4.2 378411 11 Female 101.5 2.7 2.2 276612 12 Female 99.2 5.1 4.4 519613 13 Female 99.6 3.8 3.9 392814 14 Female 99.1 2.7 3.1 277915 15 Female 99.8 4.0 4.8 408616 16 Female 100.4 4.2 2.8 433817 17 Female 99.3 4.1 2.9 417518 18 Female 100.8 3.2 2.5 329519 19 Female 98.8 5.0 2.8 511120 20 Female 99.0 3.9 4.4 399021 21 Female 101.4 2.9 3.9 297822 22 Female 99.0 1.3 4.5 138723 23 Female 100.4 4.6 3.1 472624 24 Female 99.6 3.5 3.9 355625 25 Female 100.4 3.5 3.7 358426 26 Female 101.7 4.4 2.1 454627 27 Female 101.6 4.3 5.3 442128 28 Female 99.7 4.1 3.6 419329 29 Female 97.7 4.4 4.3 452030 30 Female 102.5 4.3 4.5 438131 31 Female 100.7 3.6 3.4 367432 32 Female 100.5 1.5 3.2 161033 33 Female 100.0 4.1 4.4 422034 34 Female 100.5 3.4 2.5 354435 35 Female 99.8 3.3 5.5 344536 36 Female 100.4 2.0 3.1 212937 37 Female 99.6 3.0 5.2 312138 38 Female 98.6 3.9 5.0 401939 39 Female 101.0 4.9 3.6 495840 40 Female 101.5 3.4 4.1 349741 41 Female 99.7 3.9 2.5 398842 42 Female 98.7 3.4 3.8 354743 43 Female 100.6 2.1 4.5 222344 44 Female 100.0 3.1 3.6 318545 45 Female 98.3 3.1 3.0 320546 46 Female 100.0 3.4 2.8 354047 47 Female 99.4 4.6 3.5 470048 48 Female 99.7 4.3 4.6 436349 49 Female 98.8 3.3 3.0 343650 50 Female 101.8 3.2 3.4 334551 51 Female 99.7 4.2 2.2 429752 52 Female 98.4 4.1 4.0 415553 53 Female 100.2 2.8 4.8 291154 54 Female 100.3 2.8 5.0 289255 55 Male 99.0 3.4 4.3 514656 56 Male 97.1 3.8 1.6 575357 57 Male 99.4 2.9 4.0 443058 58 Male 100.6 3.9 4.0 592359 59 Male 99.9 3.4 3.1 519560 60 Male 99.9 2.4 3.7 368161 61 Male 100.6 3.3 2.6 511162 62 Male 98.8 1.9 4.2 290563 63 Male 101.1 4.4 3.2 675264 64 Male 100.0 5.0 1.9 757165 65 Male 100.7 2.6 3.1 404866 66 Male 101.0 2.0 4.9 303267 67 Male 100.2 3.6 3.2 545568 68 Male 99.1 2.9 4.2 439769 69 Male 101.2 5.4 4.4 820270 70 Male 98.0 3.0 3.5 454071 71 Male 99.5 3.7 3.1 563372 72 Male 99.7 3.0 3.0 464173 73 Male 99.8 2.3 4.2 348674 74 Male 101.0 3.2 2.4 488375 75 Male 100.1 1.2 3.7 189176 76 Male 100.4 4.5 3.2 680077 77 Male 99.9 3.2 1.2 483078 78 Male 99.8 5.2 2.1 785979 79 Male 100.7 3.5 4.4 531480 80 Male 101.1 2.3 3.3 353681 81 Male 97.6 3.6 4.3 551582 82 Male 100.6 2.1 5.4 320183 83 Male 100.4 1.7 5.0 271984 84 Male 99.6 3.3 4.2 503685 85 Male 101.0 2.6 3.9 393586 86 Male 99.6 3.0 3.3 460287 87 Male 99.7 3.1 5.1 471488 88 Male 100.9 2.4 4.1 371689 89 Male 101.7 2.4 2.3 374790 90 Male 100.3 2.9 3.3 439791 91 Male 99.6 4.2 1.6 636792 92 Male 98.8 1.5 3.3 231593 93 Male 99.7 3.6 0.9 549294 94 Male 99.1 3.3 4.8 510295 95 Male 99.7 4.1 2.9 619696 96 Male 100.4 2.7 3.1 414597 97 Male 99.1 3.4 3.3 515498 98 Male 102.6 3.3 4.1 500299 99 Male 100.2 2.5 4.2 3786100 100 Male 101.1 4.2 4.1 6410What is Statistics?
The science of collecting, analyzing, presenting, and interpreting data.
Myth-busting Statistics
Myth-busting Statistics
Popular Myths (false beliefs) about statistics
Myth 1
Statistics is a boring subject.
Myth 1
Statistics is a boring subject.
Statistics is a very interesting and useful subject with so many applications.
Your Turn
Assignment 1
Application of statistics in your field.
Method of evaluation: 1 slide
Time: 3 minutes
Upload your video recording to the LMS
Marks: 10 marks
Myth 2
Statistics hasn't changed much in years. It is just the same old stuff.
Myth 2
Statistics hasn't changed much in years. It is just the same old stuff.
The field of statistics has evolved significantly over time. It has also become a main componenet of many other disciplines.
Ross Ihaka and Robert Gentleman

Originators of the R programming language
R is a free software environment for statistical computing and graphics
Hadley Wickham: Chief Scientist at RStudio

- International COPSS Presidents' Award in 2019 for "influential work in statistical computing, visualisation, graphics, and data analysis"
Robert Tibshirani

- Professor in Statistics and Biomedical Data Science at Stanford University
- Received the COPSS Presidents' Award in 1996
Trevor Hastie

- Hastie is known for his contributions to applied statistics, especially in the field of machine learning, data mining, and bioinformatics.
Andrew Ng

Undergraduate degree with a triple major in computer science, statistics, and economics
Co-founded and led Google Brain, Coursera and deeplearning.ai
Data Science
Data Science
Data Science
Statistics
- Descriptive statistics/ Exploratory Data Analysis (EDA)
Use of graphical and numerical summaries to highlight the key features of data.
- Inferential statistics
Techniques for drawing conclusions about a population by examining random samples
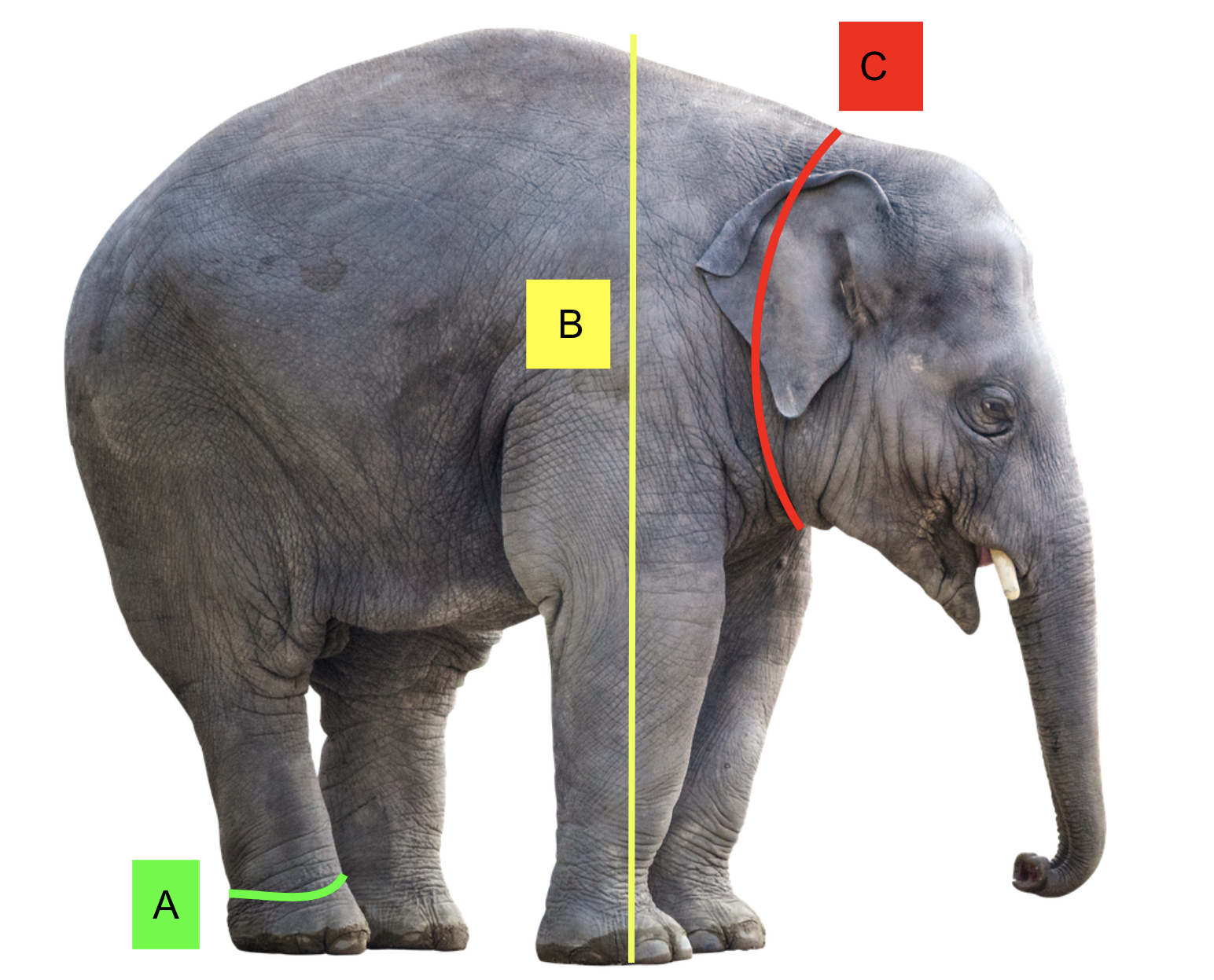
Example
Objective: Design a new chair for the university lecture halls
Wants to identify right handed left handed count
Population
A population is a complete collection of individuals/ objects that we are interested in.
Sample
A sample is a subset of a population.
Parameter
A parameter is a descriptive measure(numerical value) of the population. Parameters are usually denoted by Greek letters.
θ - population proportion
μ - population mean
Statistic
A statistic is a descriptive measure of a sample. For example, sample mean, sample standard deviation, etc. We will talk about the notations under estimator and estimate.
Variables
1. Qualitative/ Categorical
- Nominal, Ordinal
2. Quantitative/ Numerical
- Discrete, Continuous
Your turn
data("mtcars")mtcars## mpg cyl disp hp drat wt qsec vs am gear carb## Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4## Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4## Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1## Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1## Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2## Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1## Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4## Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2## Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2## Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4## Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4## Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3## Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3## Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3## Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4## Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4## Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4## Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1## Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2## Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1## Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1## Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2## AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2## Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4## Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2## Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1## Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2## Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2## Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4## Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6## Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8## Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2Your turn
data("mtcars")mtcars## mpg cyl disp hp drat wt qsec vs am gear carb## Mazda RX4 21.0 6 160.0 110 3.90 2.620 16.46 0 1 4 4## Mazda RX4 Wag 21.0 6 160.0 110 3.90 2.875 17.02 0 1 4 4## Datsun 710 22.8 4 108.0 93 3.85 2.320 18.61 1 1 4 1## Hornet 4 Drive 21.4 6 258.0 110 3.08 3.215 19.44 1 0 3 1## Hornet Sportabout 18.7 8 360.0 175 3.15 3.440 17.02 0 0 3 2## Valiant 18.1 6 225.0 105 2.76 3.460 20.22 1 0 3 1## Duster 360 14.3 8 360.0 245 3.21 3.570 15.84 0 0 3 4## Merc 240D 24.4 4 146.7 62 3.69 3.190 20.00 1 0 4 2## Merc 230 22.8 4 140.8 95 3.92 3.150 22.90 1 0 4 2## Merc 280 19.2 6 167.6 123 3.92 3.440 18.30 1 0 4 4## Merc 280C 17.8 6 167.6 123 3.92 3.440 18.90 1 0 4 4## Merc 450SE 16.4 8 275.8 180 3.07 4.070 17.40 0 0 3 3## Merc 450SL 17.3 8 275.8 180 3.07 3.730 17.60 0 0 3 3## Merc 450SLC 15.2 8 275.8 180 3.07 3.780 18.00 0 0 3 3## Cadillac Fleetwood 10.4 8 472.0 205 2.93 5.250 17.98 0 0 3 4## Lincoln Continental 10.4 8 460.0 215 3.00 5.424 17.82 0 0 3 4## Chrysler Imperial 14.7 8 440.0 230 3.23 5.345 17.42 0 0 3 4## Fiat 128 32.4 4 78.7 66 4.08 2.200 19.47 1 1 4 1## Honda Civic 30.4 4 75.7 52 4.93 1.615 18.52 1 1 4 2## Toyota Corolla 33.9 4 71.1 65 4.22 1.835 19.90 1 1 4 1## Toyota Corona 21.5 4 120.1 97 3.70 2.465 20.01 1 0 3 1## Dodge Challenger 15.5 8 318.0 150 2.76 3.520 16.87 0 0 3 2## AMC Javelin 15.2 8 304.0 150 3.15 3.435 17.30 0 0 3 2## Camaro Z28 13.3 8 350.0 245 3.73 3.840 15.41 0 0 3 4## Pontiac Firebird 19.2 8 400.0 175 3.08 3.845 17.05 0 0 3 2## Fiat X1-9 27.3 4 79.0 66 4.08 1.935 18.90 1 1 4 1## Porsche 914-2 26.0 4 120.3 91 4.43 2.140 16.70 0 1 5 2## Lotus Europa 30.4 4 95.1 113 3.77 1.513 16.90 1 1 5 2## Ford Pantera L 15.8 8 351.0 264 4.22 3.170 14.50 0 1 5 4## Ferrari Dino 19.7 6 145.0 175 3.62 2.770 15.50 0 1 5 6## Maserati Bora 15.0 8 301.0 335 3.54 3.570 14.60 0 1 5 8## Volvo 142E 21.4 4 121.0 109 4.11 2.780 18.60 1 1 4 2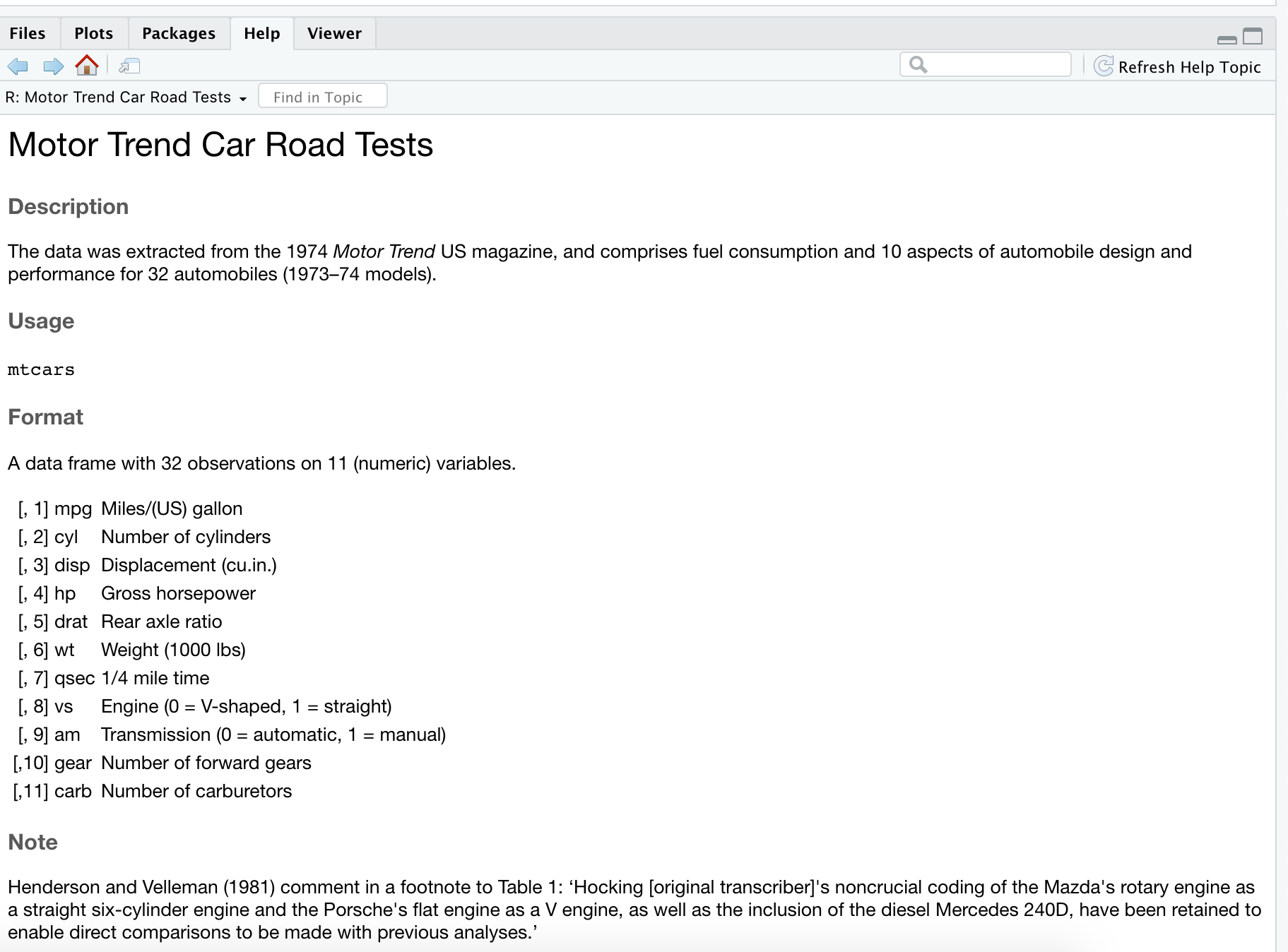
Descriptive Statistics
Data visualization: Graphics
Numerical measures
- We are using R software for data analysis and modeling
What is R?
R is a software environment for statistical computing and graphics
Language designers: Ross Ihaka and Robert Gentleman at the University of Auckland, New Zealand
Parent language: S
The latest R version 3.6.2 has been released on 2019-12-12
Why R?
Free
Powerful: Over 18000 contributed packages on the main repository (CRAN), as of July 2022, provided by top international researchers and programmers
Flexible: It is a language, and thus allows you to create your own solutions
Community: Large global community friendly and helpful, lots of resources
Numerical summary measures
summary(mtcars)## mpg cyl disp hp ## Min. :10.40 Min. :4.000 Min. : 71.1 Min. : 52.0 ## 1st Qu.:15.43 1st Qu.:4.000 1st Qu.:120.8 1st Qu.: 96.5 ## Median :19.20 Median :6.000 Median :196.3 Median :123.0 ## Mean :20.09 Mean :6.188 Mean :230.7 Mean :146.7 ## 3rd Qu.:22.80 3rd Qu.:8.000 3rd Qu.:326.0 3rd Qu.:180.0 ## Max. :33.90 Max. :8.000 Max. :472.0 Max. :335.0 ## drat wt qsec vs ## Min. :2.760 Min. :1.513 Min. :14.50 Min. :0.0000 ## 1st Qu.:3.080 1st Qu.:2.581 1st Qu.:16.89 1st Qu.:0.0000 ## Median :3.695 Median :3.325 Median :17.71 Median :0.0000 ## Mean :3.597 Mean :3.217 Mean :17.85 Mean :0.4375 ## 3rd Qu.:3.920 3rd Qu.:3.610 3rd Qu.:18.90 3rd Qu.:1.0000 ## Max. :4.930 Max. :5.424 Max. :22.90 Max. :1.0000 ## am gear carb ## Min. :0.0000 Min. :3.000 Min. :1.000 ## 1st Qu.:0.0000 1st Qu.:3.000 1st Qu.:2.000 ## Median :0.0000 Median :4.000 Median :2.000 ## Mean :0.4062 Mean :3.688 Mean :2.812 ## 3rd Qu.:1.0000 3rd Qu.:4.000 3rd Qu.:4.000 ## Max. :1.0000 Max. :5.000 Max. :8.000Numerical summary measures (cont.)
## mpg cyl disp hp ## Min. :10.40 Min. :4.000 Min. : 71.1 Min. : 52.0 ## 1st Qu.:15.43 1st Qu.:4.000 1st Qu.:120.8 1st Qu.: 96.5 ## Median :19.20 Median :6.000 Median :196.3 Median :123.0 ## Mean :20.09 Mean :6.188 Mean :230.7 Mean :146.7 ## 3rd Qu.:22.80 3rd Qu.:8.000 3rd Qu.:326.0 3rd Qu.:180.0 ## Max. :33.90 Max. :8.000 Max. :472.0 Max. :335.0 ## drat wt qsec vs ## Min. :2.760 Min. :1.513 Min. :14.50 Min. :0.0000 ## 1st Qu.:3.080 1st Qu.:2.581 1st Qu.:16.89 1st Qu.:0.0000 ## Median :3.695 Median :3.325 Median :17.71 Median :0.0000 ## Mean :3.597 Mean :3.217 Mean :17.85 Mean :0.4375 ## 3rd Qu.:3.920 3rd Qu.:3.610 3rd Qu.:18.90 3rd Qu.:1.0000 ## Max. :4.930 Max. :5.424 Max. :22.90 Max. :1.0000 ## am gear carb ## Min. :0.0000 Min. :3.000 Min. :1.000 ## 1st Qu.:0.0000 1st Qu.:3.000 1st Qu.:2.000 ## Median :0.0000 Median :4.000 Median :2.000 ## Mean :0.4062 Mean :3.688 Mean :2.812 ## 3rd Qu.:1.0000 3rd Qu.:4.000 3rd Qu.:4.000 ## Max. :1.0000 Max. :5.000 Max. :8.000Measures of central tendancy
Mean
Median
Mode
Measures of dispersion
Range
Inter quartile range
Variance
Standard deviation
R environment
The RStudio IDE
"If R were an airplane, RStudio would be the airport, providing many, many supporting services that make it easier for you, the pilot, to take off and go to awesome places. Sure, you can fly an airplane without an airport, but having those runways and supporting infrastructure is a game-changer."
-- Julie Lowndes
Create a new project
R Console
7+1[1] 8rnorm(10) [1] -0.572542604 -1.363291256 -0.388722244 0.277914132 -0.823081122 [6] -0.068840934 -1.167662326 -0.008309014 0.128855402 -0.145875628Variable assignment
a <- rnorm(10)a [1] -0.16391096 1.76355200 0.76258651 1.11143108 -0.92320695 0.16434184 [7] 1.15482519 -0.05652142 -2.12936065 0.34484576Variable assignment
a <- rnorm(10)a [1] -0.16391096 1.76355200 0.76258651 1.11143108 -0.92320695 0.16434184 [7] 1.15482519 -0.05652142 -2.12936065 0.34484576b <- a*100b [1] -16.391096 176.355200 76.258651 111.143108 -92.320695 16.434184 [7] 115.482519 -5.652142 -212.936065 34.484576Data permanency
ls()can be used to display the names of the objects which are currently stored within R.The collection of objects currently stored is called the workspace
ls() [1] "a" "A" "a1" "a2" "b" "B" "b1" "b2" [9] "b3" "C" "c1" "c2" "df" "g1" "Gender" "ID" [17] "mtcars" "p1" "w1" "w2" "w3" "Weight"To remove objects the function
rmis availableremove all objects
rm(list=ls())remove specific objects
rm(x, y, z)
rm(a)ls() [1] "A" "a1" "a2" "b" "B" "b1" "b2" "b3" [9] "C" "c1" "c2" "df" "g1" "Gender" "ID" "mtcars"[17] "p1" "w1" "w2" "w3" "Weight"rm(list=ls())ls()character(0)At the end of an R session, if save: the objects are written to a file called .RData in the current directory, and the command lines used in the session are saved to a file called .Rhistory
When R is started at later time from the same directory
When R is started at later time from the same directory it reloads the associated workspace and commands history.
When R is started at later time from the same directory it reloads the associated workspace and commands history.
Comment your code
- Each line of a comment should begin with the comment symbol and a single space: # .
rnorm(10) # This is a comment [1] -1.9049554 -0.8111702 1.3240043 0.6156368 1.0916690 0.3066049 [7] -0.1101588 -0.9243128 1.5929138 0.0450106sum(1:10) # 1+2[1] 55Style Guide
- Good coding style is like correct punctuation: you can manage without it, butitsuremakesthingseasiertoread. -- Hadley Wickham
sum(1:10)#Bad commenting style[1] 55sum(1:10) # Good commenting style[1] 55- Also, use commented lines of - and = to break up your file into easily readable sub-sections.
# Read data ----------------# Plot data ----------------To learn more read Hadley Wickham's Style guide.
Objects in R
- R is an object-oriented language.
- An object in R is anything (data structures, functions, etc., that can be assigned to a variable).
Let's take a look of some common types of objects.
Let's take a look of some common types of objects.
1. Data structures are the ways of arranging data.
- You can create objects, using the left pointing arrow <-
2. Functions tell R to do something.
A function may be applied to an object.
Result of applying a function is usually an object too.
- All function calls need to be followed by parentheses.
Example
a <- 1:20 # data structuresum(a) # sum is a function applied on a[1] 210help.start() # Some functions work on their own.Getting help with functions and features
- R has inbuilt help facility
Method 1
help(rnorm)- For a feature specified by special characters such as
for,if,[[
help("[[")- Search the help files for a word or phrase.
help.search(‘weighted mean’)Method 2
?rnorm??rnormData structures
Data structures differ in terms of,
Type of data they can hold
How they are created
Structural complexity
Syntax to identify and access individual elements
1. Vectors
Vectors
Vectors are one-dimensional arrays that can hold numeric data, character data, or logical data.
Combine function c() is used to form the vector.
Data in a vector must only be one type or mode (numeric, character, or logical). You can’t mix modes in the same vector.
Vector assignment
Syntax
vector_name <- c(element1, element2, element3)x <- c(5, 6, 3, 1 , 100)assignment operator ('<-'), '=' can be used as an alternative.
c()function
What will be the output of the following code?
y <- c(x, 500, 600)Types and tests with vectors
first_vec <- c(10, 20, 50, 70)second_vec <- c("Jan", "Feb", "March", "April")third_vec <- c(TRUE, FALSE, TRUE, TRUE)fourth_vec <- c(10L, 20L, 50L, 70L)To check if it is a
- vector:
is.vector()
is.vector(first_vec)[1] TRUE- charactor vector:
is.charactor()
is.character(first_vec)[1] FALSE- double:
is.double()
is.double(first_vec)[1] TRUE- integer:
is.integer()
is.integer(first_vec)[1] FALSE- integer:
is.integer()
is.integer(first_vec)[1] FALSE- logical:
is.logical()
is.logical(first_vec)[1] FALSE- length
length(first_vec)[1] 4Coercion
Vectors must be homogeneous. When you attempt to combine different types they will be coerced to the most flexible type so that every element in the vector is of the same type.
Order from least to most flexible
logical --> integer --> double --> character
a <- c(3.1, 2L, 3, 4, "GPA") typeof(a)[1] "character"anew <- c(3.1, 2L, 3, 4)typeof(anew)[1] "double"Explicit coercion
Vectors can be explicitly coerced from one class to another using the as.* functions, if available. For example, as.charactor, as.numeric, as.integer, and as.logical.
vec1 <- c(TRUE, FALSE, TRUE, TRUE)typeof(vec1)[1] "logical"vec2 <- as.integer(vec1)typeof(vec2)[1] "integer"vec2[1] 1 0 1 1Why does the below output NAs?
x <- c("a", "b", "c")as.numeric(x)[1] NA NA NAx1 <- 1:3x2 <- c(10, 20, 30)combinedx1x2 <- c(x1, x2)combinedx1x2[1] 1 2 3 10 20 30x1 <- 1:3x2 <- c(10, 20, 30)combinedx1x2 <- c(x1, x2)combinedx1x2[1] 1 2 3 10 20 30class(x1)[1] "integer"class(x2)[1] "numeric"class(combinedx1x2)[1] "numeric"- If you combine a numeric vector and a character vector
y1 <- c(1, 2, 3)y2 <- c("a", "b", "c")c(y1, y2)[1] "1" "2" "3" "a" "b" "c"Name elements in a vector
You can name elements in a vector in different ways. We will learn two of them.
When creating it
x1 <- c(a=1991, b=1992, c=1993)x1## a b c ## 1991 1992 1993Modifying the names of an existing vector
x2 <- c(1, 5, 10)names(x2) <- c("a", "b", "b")x2## a b b ## 1 5 10Note that the names do not have to be unique.
To remove names of a vector
Method 1
unname(x1); x1[1] 1991 1992 1993 a b c 1991 1992 1993Method 2
names(x2) <- NULL; x2[1] 1 5 10What will be the output of the following code?
v <- c(1, 2, 3)names(v) <- c("a")vSimplifying vector creation
- colon
:produce regular spaced ascending or descending sequences.
10:16[1] 10 11 12 13 14 15 16-0.5:8.5 [1] -0.5 0.5 1.5 2.5 3.5 4.5 5.5 6.5 7.5 8.5- sequence:
seq(initial_value, final_value, increment)
seq(1,11) [1] 1 2 3 4 5 6 7 8 9 10 11seq(1, 11, length.out=5)[1] 1.0 3.5 6.0 8.5 11.0seq(0, 11, by=2)[1] 0 2 4 6 8 10- repeats
rep()
rep(9, 5)[1] 9 9 9 9 9rep(1:4, 2)[1] 1 2 3 4 1 2 3 4rep(1:4, each=2) # each element is repeated twice[1] 1 1 2 2 3 3 4 4rep(1:4, times=2) # whole sequence is repeated twice[1] 1 2 3 4 1 2 3 4rep(1:4, each=2, times=3) [1] 1 1 2 2 3 3 4 4 1 1 2 2 3 3 4 4 1 1 2 2 3 3 4 4rep(1:4, 1:4) [1] 1 2 2 3 3 3 4 4 4 4rep(1:4, c(4, 1, 4, 2)) [1] 1 1 1 1 2 3 3 3 3 4 4Logical operators
c(1, 2, 3) == c(10, 20, 3)[1] FALSE FALSE TRUEc(1, 2, 3) != c(10, 20, 3)[1] TRUE TRUE FALSE1:5 > 3[1] FALSE FALSE FALSE TRUE TRUE1:5 < 3[1] TRUE TRUE FALSE FALSE FALSE<=less than or equal to>=greater than or equal to|or&and
Operators: %in% - in the set
a <- c(1, 2, 3)b <- c(1, 10, 3)a%in%b[1] TRUE FALSE TRUEx <- 1:10y <- 1:3x [1] 1 2 3 4 5 6 7 8 9 10y[1] 1 2 3x %in% y [1] TRUE TRUE TRUE FALSE FALSE FALSE FALSE FALSE FALSE FALSEy %in% x[1] TRUE TRUE TRUEVector arithmetic
- operations are perfored element by element.
c(10, 100, 100) + 2 # two is added to every element in the vector[1] 12 102 102Vector arithmetic
- operations are perfored element by element.
c(10, 100, 100) + 2 # two is added to every element in the vector[1] 12 102 102- operations between two vectors
v1 <- c(1, 2, 3); v2 <- c(10, 100, 1000)v1 + v2[1] 11 102 1003Add two vectors of unequal length
longvec <- seq(10, 100, length=10); shortvec <- c(1, 2, 3, 4, 5)shortvec + longvec [1] 11 22 33 44 55 61 72 83 94 105What will be the output of the following code?
first <- c(1, 2, 3, 4); second <- c(10, 100)first * secondReferences
Le Dinh, T., Lee, S. H., Kwon, S. G., & Kwon, K. R. (2022). COVID-19 Chest X-ray Classification and Severity Assessment Using Convolutional and Transformer Neural Networks. Applied Sciences, 12(10), 4861.